Hello everyone, just checking in to thank Steven for hosting my notebook for so long and creating such a cool wikispace.
I recently returned from Belgium, where I met many Postdocs interested in developing their knowledge and skills in molecular techniques for their particular projects. Most of the postdocs were more interested in detecting change in their ecosystems due to natural and anthropogenic disturbance. Some of the students were doing exploratory studies in extreme environments, such as Antarctic lakes or the deep sea vents off the coast of Europe. A few were aquaculturists hoping to learn how to improve their existing research with genomics techniques. Most notably, there was a gentleman from Bangladesh working on feeding nematodes to early life stages of shrimp. His work is very closely related to mine in regards to the fatty acid desaturases.
I will be providing a little summary of the genomics work soon, but I just wanted to make a note that I have developed a workspace for the students of the class called marinebiologygenomics.wikispaces.com and I wanted to thank Steven for the inspiration by developing this wikispace. It is a really great tool, and since the students of my class were so geographically dispersed, I hope that it will facilitate their educational process. I think I have a better understanding of how to describe the methods and tools of genomics research since I started from scratch about two years ago. Of course, I did get a lot of help from sitting in on Steven's class as well as taking the biotech certificate classes at Seattle Central Community College (this program is now sadly defunct, due to the slacking economy in Seattle --- hopefully they will bring it back.) And, of course I have to thank Sam White for being so patient and answering all my simple questions -- he is definitely the glue holding the lab together.
If anyone has suggestions for links or information for the students who will be using marinebiologygenomics.wikispaces.com, please let me know. Also, please feel free to join.
Adelaide
September 14, 2009
Okay, so I am not in Seattle at this time, but I am still trying to work out the kinks in this project.
I have reformatted the lab notes here so that the most recent observations will be at the top from now on.
For those of you waiting eagerly for the results of the latest sequencing, once Steven returned we realized that the latest gel was more diagnostic of a failed transformation than an actual confirmation of product. All sequences matched the vector primer, which is a beta galactosidase which is an enzyme for breaking the beta bonds in oligosaccharides, by the way.
A helpful observation from this recent experience is to add XGal to the plates used after the initial transformation. If the colonies are still white, the transformation was successful. This step is usually not necessary when there are large colonies and it is easy to distinguish whether a colony is truly white or just very very light blue, indicating that the transformation did not take place.
The additional sequences on the Tigriopus genomic DNA was also very similar to last year's sequence. No additional proof was garnered to indicate that the gene was definitely related to the copepod. It was frustrating that new PCR's on the genomic DNA from last year did not provide the same stellar results.
So, I am starting again here at Ghent University in Belgium. I will be working with a species of copepod that has demonstrated the ability to convert HUFA's, much like my original species used in my dissertation. This copepod has been verified to do the conversion by the co-PI on this project, Marleen DeTroch. She used stable isotopes to label the copepod food and was able to trace the production of DHA, 22:6n3, from shorter chain precursors. This work is still inferential in the respect that while we may assume that delta-5 and delta-6 desaturases are the pathways, they have not been definitively found in copepods.
This week, we will be going out to the field to collect the harpacticoid copepod Microarthridion littorale from an intertidal zone that borders the Netherlands. We will bring the copepod into the lab, starve them, then provide them two diets: HUFA-rich (diatom) and HUFA-deficient (bacteria). After twenty-four hours we will extract their mRNA, and do some genefishing (hopefully). In addition, we will continue to develop and test primers for our genes of interest. A recent NCBI gene published by the Norwegian sea lice group may provide a better template for the primers at this time.
In addition, we will collect genomic DNA from any copepods that we find in abundance to test the presence/absence of any lipid desaturase or elongation genes that we can.
August 13, 2009
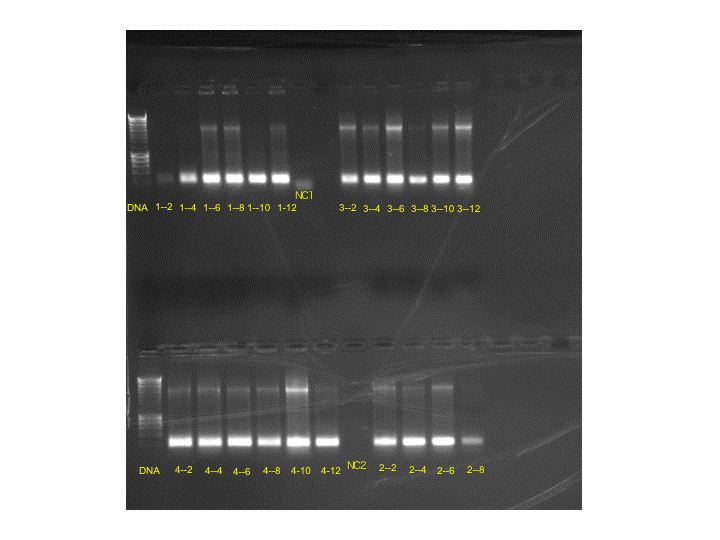
Results of the gel were very uniform, so only two bands were chosen for more grow-out in LBKan50.
Bands: 3-12 and 4-10 were chosen. Samples from these streaks were placed in LBKan50 at 37 degrees C for overnight development.
Tomorrow, miniprep.....
August 11, 2009
Okay, so I had 45 different colonies of bacteria, all very white, which means they retained their kanamyacin resistance. I prepared a PCR for all the even numbered colonies.
Plate 1: 2,4,6,8,10, 12
Plate 2: 2,4,6,8
Plate 3: 2,4,6,8,10,12
Plate 4: 2,4,6,8,10,12
PCR Reaction:
| 25 uL |
GoTaq Mixture, Note: new GoTaq mixture used, Apex TaqRED Master Mix 2.0X 1.5mM MgCL2, lot # BLM-09F30 |
| 1 uL |
M13 Forward Primer 10 uM (from -20 degrees C box) |
| 1 uL |
M13 Reverse Primer 10 uM (from -20 degrees C box) |
| 23 uL |
H20 for PCR |
| 50 uL |
TOTAL |
The PCR Reaction was set up, using the following parameters:
| Segment |
|
Temp (degrees C) |
Duration (minutes) |
|
| 1 |
1 |
94 |
5 |
|
| 2 |
40 |
94 |
1 |
|
| 3 |
40 |
55 |
1 |
|
| 4 |
40 |
72 |
1 |
|
| 5 |
1 |
72 |
10 |
|
Gel electropheresis will be conducted tomorrow......
August 10, 2009
The colonies were prepared for PCR. Unfortunately, I forgot to hit "RUN" on thermal cycler.
August 7, 2009
The bacterial colonies from the 8/4 plates which were transfered to the gridded plates showed a nice development. There are plenty of cells, and they are all white in color, indicating that they carry the kanamyacin resistance. I have approximately 45 distinct colonies to choose from.
No development occured on the last set of cells transformed (labeled 8/6). These plates were stored in the fridge.
August 6, 2009
The bacterial colonies have appeared, but are growing very slowly, they were left until the end of the day at 36 degrees C. Several white and light blue colonies as well as dark blue colonies have appeared dispersed about the two plates labeled 8/4.
Only the 8/4 cells have apparent colonies. The transformation on 8/5 apparently did not take. The leftover cells from the 8/5 transformation were spread on two plates and labeled 8/6.
Results of the PCR Reaction:
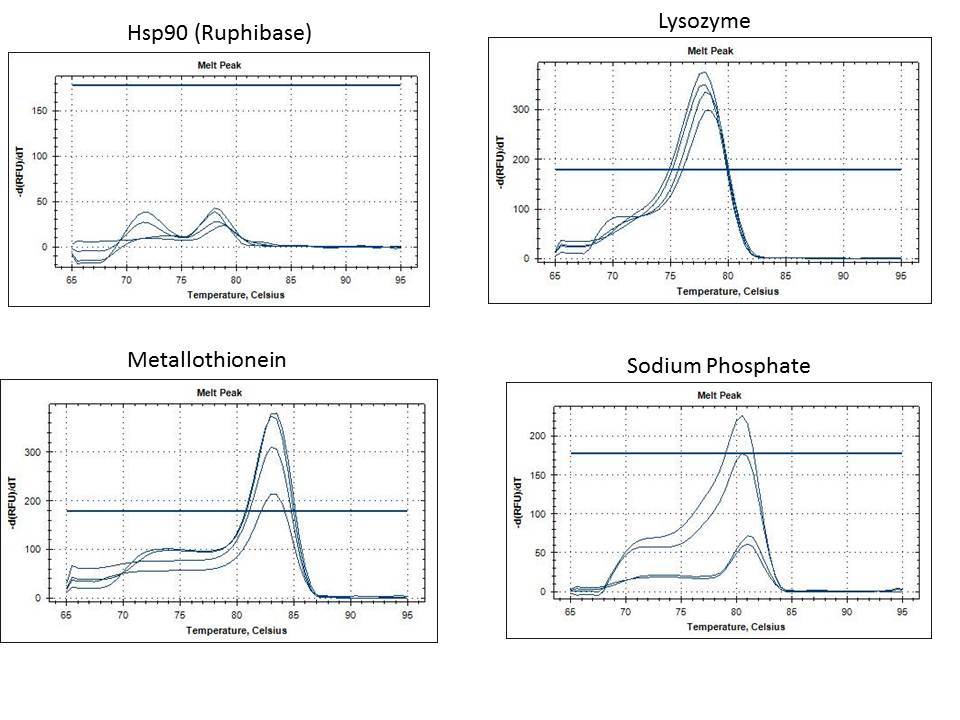
While I did not replicate the original result of bands from the genomic Tigriopus DNA, I am still getting a strong signal from the two bands isolated from last year.
I cut these two bands for cloning.
At the end of the day, I screened the 8/4 plates for colonies that were white or light blue and smeared thm onto 4 gridded plates.
August 5, 2009
.... the plates apparently had expired. They were made up about one month ago, so the antibiotic had probably expired. No distinct colonies were apparent on the two plates.
So, today, I poured some more plates with Kanamyacin instead of Ampicillin.
To make the plates, I added 1.5 mg Agar (Bacto Agar, Lot 6275177, 2011-08-31) to 100 mL 1x LB and autoclaved.
After autoclaving the agar was cooled to 55 degrees C, and 1 ug/mL Kanamyacin was added to the broth before pouring the plates.
The cells left over from the previous attempt were reintroduced to the agar. 100 uL of SOC medium was added to the leftover cells to facilitate the transfer of a larger volume to the plates (50 uL and 150 uL). (labeled 8/4)
A new transformation was done using the leftover 2 uL from the TOPO Cloning reaction (labeled 8/5).
I had some time to do some PCR, so I tried to replicate the original successful PCR product from last year.
PCR Reaction:
25 uL Taq
1 uL Forward Primer delta-6
1 uL Reverse Primer delta-6
4 uL Template
19 uL water
Templates =
genomicDNA1 from Tigriopus 2008
genomicDNA2 from Tigriopus 2008
Tigriopus Top band from 2008
Tigriopus Bottom band from 2008
Tigriopus cDNA from Isochrysis treatment 2009
Tigriopus cDNA from Tetraselmis treatment 2009
The PCR Reaction was set up as follows:
| Segment |
|
Temp (degrees C) |
Duration (minutes) |
|
| 1 |
1 |
95 |
10 |
|
| 2 |
40 |
95 |
1 |
|
| 3 |
40 |
55 |
1 |
|
| 4 |
40 |
72 |
1 |
|
| 5 |
1 |
72 |
10 |
August 4, 2009
Since the straight sequencing of PCR product did not seem to yield useful sequences, we decided to try cloning.
This protocol followed the Invitrogen TOPO TA Cloning kit and the BioLine CH3-Blue Chemically competent cells.
Taq polymerase had been utilized for the PCR products, so a single deoxyadenosine (A) had been added to the 3' ends of the PCR products. The vector in teh TOPO kit has a single overhanging 3' deoxythymidine (T) residue, which allows the PCR inserts to ligate with the vector.
TOPO Cloning Reaction setup:
| Reagent |
Chemically competent E. coli |
| Fresh PCR product |
4 uL |
| Salt Solution |
1 uL |
| Water |
add to total volume of 5 uL (not needed here) |
| TOPO vector |
1 uL |
| Final volume |
6 uL |
2. Place reaction on ice
3. If not used immediately, the TOPO reaction can be stored at -20 degrees C overnight
Transformation Procedure:
Beforehand:
Equilibrate a water bath to 42 degrees C
Warm vial of X.O.C. medium to room temperature
Warm selective plates at 37 degrees C for at least 30 minutes
Spread 40 uL of 40 mg/mL X-gal on each LB plate and incubate at 37 degrees C until use
Since we were using BioLine competent cells, we did not thaw beforehand, they were kept frozen in the -80 degrees C freezer.
Procedure:
1. Add 2 uL of the TOPO cloing reaction above into slightly thawed BioLine vial of cells.
2. Incubate on ice for 5 minutes
3. Heat-shock the cells for 30 seconds at 42 degrees C without shaking
4. Immediately tranfer the tubes to ice
5. Add 250 uL S.O.C. medium
6. Cap the tube tightly and shake the tube horizontally at 37 degrees C for one hour
7. Spread 30 50 uL of the cells onto a prewarmed plate, and 150 uL onto a second plate
8. Incubate overnight at 37 degrees C.
We used LBAMP50 plates prepared early July, 2008.
So, we set it up for overnight, and....
July 31, 2009
>BAND DD1: 043_F12_D1 Trace of /Users/srlab/Desktop/Run_sam/043_F12_D1.ab1; length: 442; ambiguous 346; low quality 392; medium quality 44; high quality 6
NNNNNNNNGNNNNNNNNNCNGANNNNNTGGNCGAGAAGGGANAGAACNTCTTGANCATTC
CCNNNANTGNCNNCCNTNNNNNNNNNNNNNNNNGNNNNNNNNNGNNNANGNNNNNNNNNN
ANGGNGTNNNNNNTTTNNNNNNCNNNTNTNNCCNNNNNNNNNNNNNCNNNNNNNNNNNNN
NNCNNNNNNNNNNNNNCNNNNNNNNNNNNNNNNNNNNNANNNNNCNNNNNNNNNNNNNNN
NNCCCNNNNANNNNTTNGNNNNNNANANNNNTTNNTTNNNANNNNNNNANNNNNNNNNNN
NNTNNNNNNNNTNNNNTNNNNNNNNNNNNNNNNNNNNNNNNNNNNAANNNNNNNNNNNNN
NNGNNNNNNNNCNNNTCNNTNNNNNNNNNNNNNNNCNTNNNNNNNNNTNCNNNNNNTNNN
NNNNNNNNNNNNTNNTNNNNNN
>BAND DD6: 044_E12_D6 Trace of /Users/srlab/Desktop/Run_sam/044_E12_D6.ab1; length: 229; ambiguous 190; low quality 217; medium quality 12; high quality 0
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNAACNNNNNNNNNNNNNCGNNNNGN
NNGNNGCNNNNCCNTNNNNNNNNNNNNNNNNNNNNCCNNNNTTNNTNNNNNNNNNNNNNN
NNCNNNNNNNNNNNTNNNNNTTTNNNNNTTNNNNNNNNTNCTNNNNNNNNNNNTNTNNNT
NNTNNNCNCNNNNNANNNNNNNNNNTNNNNNNCNNNNNCCCNNNNNNNN
>BAND DD11: 045_D12_D11 Trace of /Users/srlab/Desktop/Run_sam/045_D12_D11.ab1; length: 398; ambiguous 84; low quality 103; medium quality 73; high quality 222
NNNNNNNNNNNTTTGNNGGANCTTNNTCGAAGAGTNTGGACCTGATATCATCGACCAACT
CGTCAACGAGAACCTCTCGCCCAGTCTCCTTTGCGGACCCCAGGGCTTGGGAATTTGCGA
TGCTTAAAACTGATCTTTGGATCAAGAATTGAGACGCGTTCGTGTGGAAATTCTTGCCAT
AATGATAGTTGTCATCCAAATATTAGTGATCGAACGGTTTATTCATTCCACAAACTCTAA
AGGATTGAAAAACTTTGAGTCAATTTCCCATTTCTGGGTAAATAAAGTGATTTGGTAAAT
ANNANAAAAAAAAAAAAAANCNNNNNNTGGGGNGGNNNNNNNNNNNNNNNNNNNNNNNNN
NNNNNNNNNNNNNNGGGGNNNNNNNNNNNNCCNNCNNN
>BAND DD16 046_C12_D16 Trace of /Users/srlab/Desktop/Run_sam/046_C12_D16.ab1; length: 241; ambiguous 165; low quality 197; medium quality 43; high quality 1
NNNNNNNNNNNNNNNNNNNNNNNNNNNNATNNGNTTTCTCCTTGAANNNNNNNTTNGNAG
NCGGACGACCCGGATNNGTNNNNANNANNAANGNCNAGANGGGAGGNNATNCNAANNNNN
NNNNNNNNANNNNNNNNNGNNNNNNNANTNNNNNNANGNNGGNNNNNNNNNNNNCNNNNN
NNNNNCNNNNTNNNNNGNNNCNNNNCNNNNNNCNANNNTNNCNNNNNCNNNNNNNNNCNN
N
>Acartia delta-6 band with forward primer: 047_B12_A2F Trace of /Users/srlab/Desktop/Run_sam/047_B12_A2F.ab1; length: 348; ambiguous 59; low quality 95; medium quality 172; high quality 81
GNNNNNNNNNNNNNNGNNNNNNGNNNNNCANATGGCTGANCTTGTTNNNNNNCCTCTGCT
GATCANATGTTTGGAGACAGTTATTTCTCCATATTTNCCCCCCTCACCCCTGCCATAGTG
ACCTCTGGGGTTGNAGCTTANCNTGGACAGTTCAGCATCAGGGTTGATATTGACAATGTC
TACCCCNGTCAGGGAGTAGCTGGTCTTGTCCAATCTGTACATGAGGATGCTGATATGAGA
GACAATGTGGACTTTANCGCAAGTGGTGNANNNNNNGATAGTTNNNNNNNGTGATATTAG
GTTGGATAAACCAAAGGTTNGTTGNGTTATGCTTGTCCTTCCACCNNN
>Calanus delta-6 band with forward primer: 048_A12_C1F Trace of /Users/srlab/Desktop/Run_sam/048_A12_C1F.ab1; length: 422; ambiguous 173; low quality 204; medium quality 178; high quality 40
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNANNNNNNNNNN
NNNNNNNGANNNNNNCCNNNNNNNNNAANTTNNCNNNNTCACCCCTGCCATAGTGACCTC
TGGGGTTGTAGCTTANCATGGACAGTTCAGCATCAGGGTTGATAGTGACAATGTCTACCC
CAGTCAGGGAGTANCTGGTCTTGTCCAATCTGTACATGAGGATGCTGATATGANANNNNN
TGTGNACTTTANNGNAAGTGNTGNAGGTGGNGATAGTTATGCTCANNGATATTANGTTGG
ATNNNNNNNAGGTTTGTTGGGTTATGCTTGTCCTTCCACNNNNNNNNNNNNNCNNCCCNN
NNNNGGNTNGNCNNNNNCNGNNANTCCTTCCACNNNNNNNTANNNNNNNNNNNNNNNNNN
NN
While there appeared to be many contiguous sequences, there were no hits in the databases on any of them.
July 17, 2009
A few promising bands from the previous gels were picked out for sequencing, to make sure that zooplankton RNA were extracted preferentially over any possible phytoplankton RNA from the diets.
First, a spectrophotometric analysis of DNA extracted from the agarose using the DNA quick prep kit was conducted:
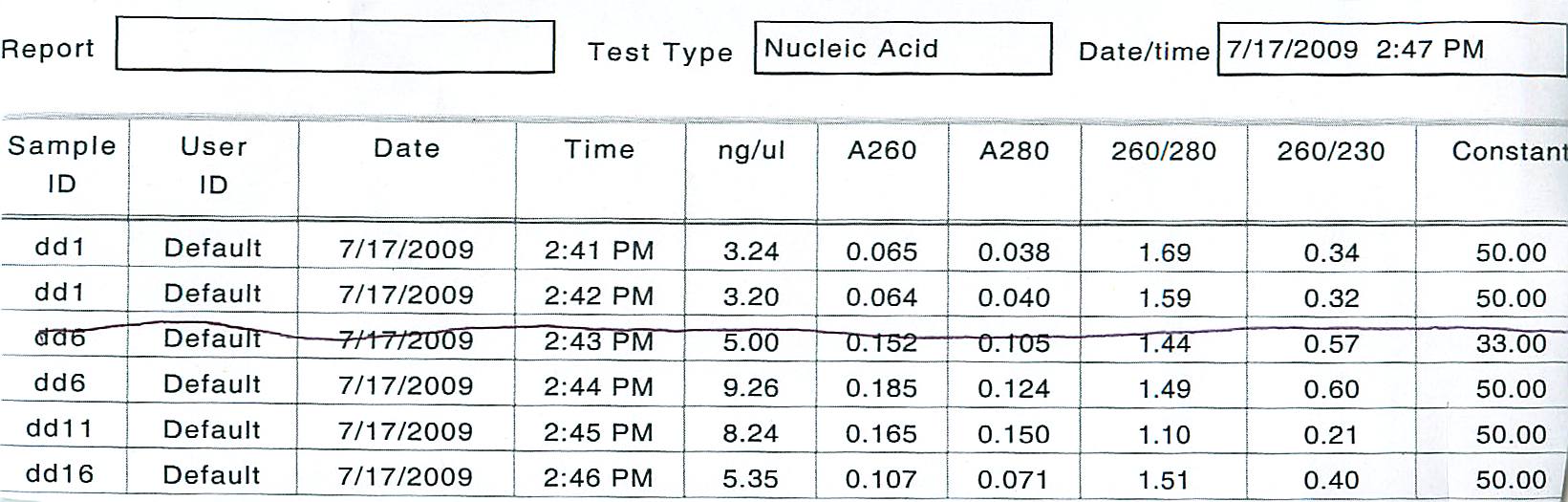
The DNA was prepared for analysis using the following amounts.
| Sample ID |
DNA concentration (ng/uL) |
Volume of DNA (uL) |
Primer Added |
DIH2O |
Final Volume |
Final Concentration |
| DD1 |
3.25 |
14 uL |
2 uL ACP9 |
16 uL |
2.84 ng/uL |
|
| DD6 |
9.26 |
10 uL |
2 uL ACP17 |
12 uL |
7.72 ng/uL |
|
| DD11 |
8.24 |
5.4 uL |
2 uL ACP42 |
2.6 uL |
10 uL |
4.45 ng/uL |
| DD16 |
5.35 |
10 uL |
2 uL ACP48 |
2 uL |
14 uL |
3.82 ng/uL |
| A2 |
14,93 |
3 uL |
1 uL Delta-6 Forward |
2 uL |
6 uL |
7.47 ng/uL |
| C1 |
11.77 |
5 uL |
1 uL Delta-6 Forward |
6 uL |
9.81 ng/uL |
These samples will be sent for sequencing analysis.
July 16, 2009
Okay, after having a really excellent result on the first two gels, I am now humbled, because I had a terrible third gel. I had some trouble loading the samples, and the bands were quite inconsistent and the picture was difficult to take because of the dye on the gel. Here is the result, I did still get some bands:
PCR results from 3rd Set of reactions (GEL 3):
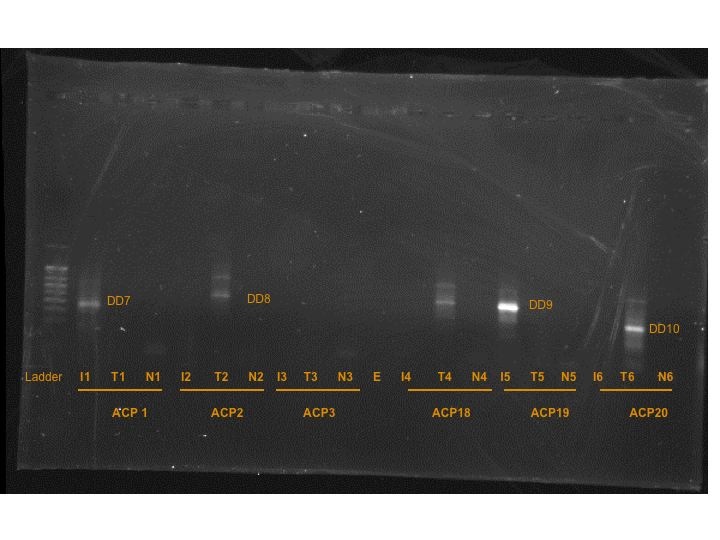
I did cut out the bands that were present, but they wouldn't be truly a set of "differentially expressed" bands because many of them did not have matches, like in the previous two gels.
DD7 = band from T-Iso cDNA x ACP1
DD8 = band from Tetraselmis cDNA x ACP2
DD9 = band from T-Iso cDNA x ACP19
DD10 = band from Tetraselmis cDNA x ACP20
Another set of PCR reactions was set up with a different set of primers:
| ACP Random Primer |
T-Iso Treated copepod cDNA |
Tetraselmis Treated copepod cDNA |
Control |
GEL |
|||
| ACP41 |
IA1 |
TA1 |
NA1 |
4 |
|||
| ACP42 |
IA2 |
TA2 |
NA2 |
4 |
|||
| ACP43 |
IA3 |
TA3 |
NA3 |
4 |
|||
| ACP44 |
IA4 |
TA4 |
NA4 |
4 |
|||
| ACP45 |
IA5 |
TA5 |
NA5 |
4 |
|||
| ACP46 |
IA6 |
TA6 |
NA6 |
5 |
|||
| ACP53* |
IA7 |
TA7 |
NA7 |
5 |
|||
| ACP48 |
IA8 |
TA8 |
NA8 |
5 |
|||
| ACP49 |
IA9 |
TA9 |
NA9 |
5 |
|||
| ACP52 |
IA10 |
TA10 |
NA10 |
5 |
|||
To simplify gel loading and band cutting, two 16 well gels were used for these 30 reactions.
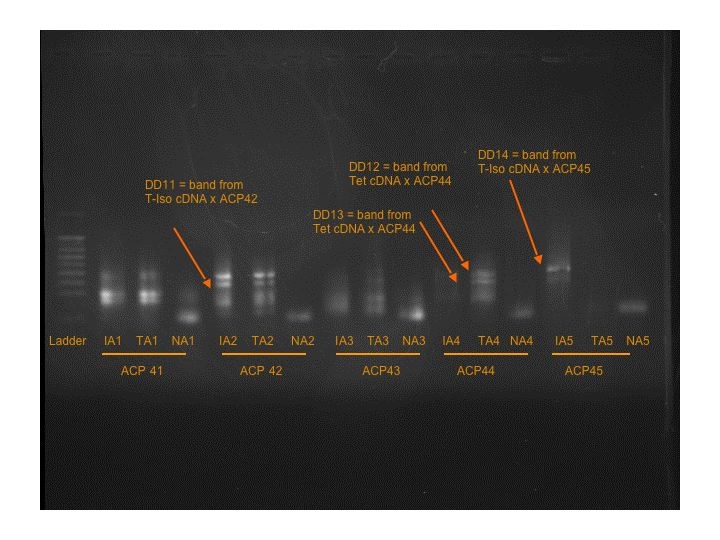
Seven more bands were found on Gels 4 and 5.
T-Iso cDNA x ACP42: DD11
Tetraselmis cDNA x ACP44: DD12 and DD13
T-Iso cDNA x ACP45: DD14
T-Iso cDNA x ACP53: DD15
Tetraselmis cDNA x ACP48: DD16 ----- strange that there is a band in the control, possible pipetting contamination upon loading.
Tetraselmis cDNA x ACP52: DD17
July 15, 2009
An 0.8% agarose gel was prepared for the PCR products.
Products were loaded in the following order:
DNA ladder, ATI1, ATT1, ANC1, empty, ATI2, ATT2, ANC2, empty, ATI3, ATT3, ANC3, empty, ATI4, ATT4, ANC4
For some reason, the ATI3 PCR product (ACP9xT-Iso treatment) did not have the same volume as the other PCR products.
Here are the results of the 1st set of PCR reactions (GEL 1):
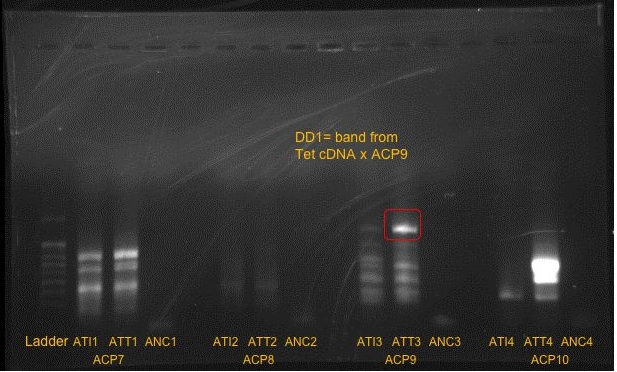
The band furthest to the right was a little indistinguishable, so a closeup photo was taken:
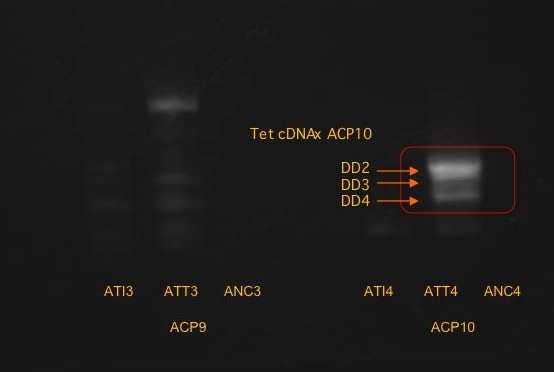
It was possible to distinguish the three bands on the right.
Samples to the right indicated copepods fed Tetraselmis.
It is possible that the difference seen in the third reaction from the left was due to the smaller amount of PCR product loaded into the gel.
One band was isolated from the third reaction (ACP9 x Tetraselmis treatment): DD1
Three bands were cut and isolated from the fourth reaction (ACP10 x Isochrysis): DD2, DD3, DD4 from top to bottom.
Two more sets of PCR reactions were prepared with half the cDNA template due to the smearing from the first set of reactions.
| T-ISO |
T-TET |
DIH2O |
||
| cDNA |
1 uL |
1.45 uL |
0 uL |
|
| arbitraty ACP 9 #7 through #10 |
2 uL |
2 uL |
2 uL |
|
| 10 uM dT-ACP-2 |
1 uL |
1 uL |
1 uL |
|
| 2 x SeeAMP ACP Master Mix |
10 uL |
10 uL |
10 uL |
|
| DIH2O |
6 uL |
5.55 uL |
7 uL |
|
| TOTAL |
20 uL |
20 uL |
20 uL |
|
| ACP Random Primer |
T-Iso Treated copepod cDNA |
TetraselmisTreated copepod cDNA |
Control |
GEL |
|
| ACP7 |
ATI1 |
ATT1 |
ANC1 |
1 |
|
| ACP8 |
ATI2 |
ATT2 |
ANC2 |
1 |
|
| ACP9 |
ATI3 |
ATT3 |
ANC3 |
1 |
|
| ACP10 |
ATI4 |
ATT4 |
ANC4 |
1 |
|
| ACP12 |
AI1 |
AT1 |
NC1 |
2 |
|
| ACP13 |
AI2 |
AT2 |
NC2 |
2 |
|
| ACP14 |
AI3 |
AT3 |
NC3 |
2 |
|
| ACP5 (no 15 left) |
AI4 |
AT4 |
NC4 |
2 |
|
| ACP16 |
AI5 |
AT5 |
NC5 |
2 |
|
| ACP17 |
AI6 |
AT6 |
NC6 |
2 |
|
| ACP1 |
I1 |
T1 |
N1 |
3 |
|
| ACP2 |
I2 |
T2 |
N2 |
3 |
|
| ACP3 |
I3 |
T3 |
N3 |
3 |
|
| ACP18 |
I4 |
T4 |
N4 |
3 |
|
| ACP19 |
I5 |
T5 |
N5 |
3 |
|
| ACP20 |
I6 |
T6 |
N6 |
3 |
|
Here are the results from the second set of PCR reactions (GEL 2):
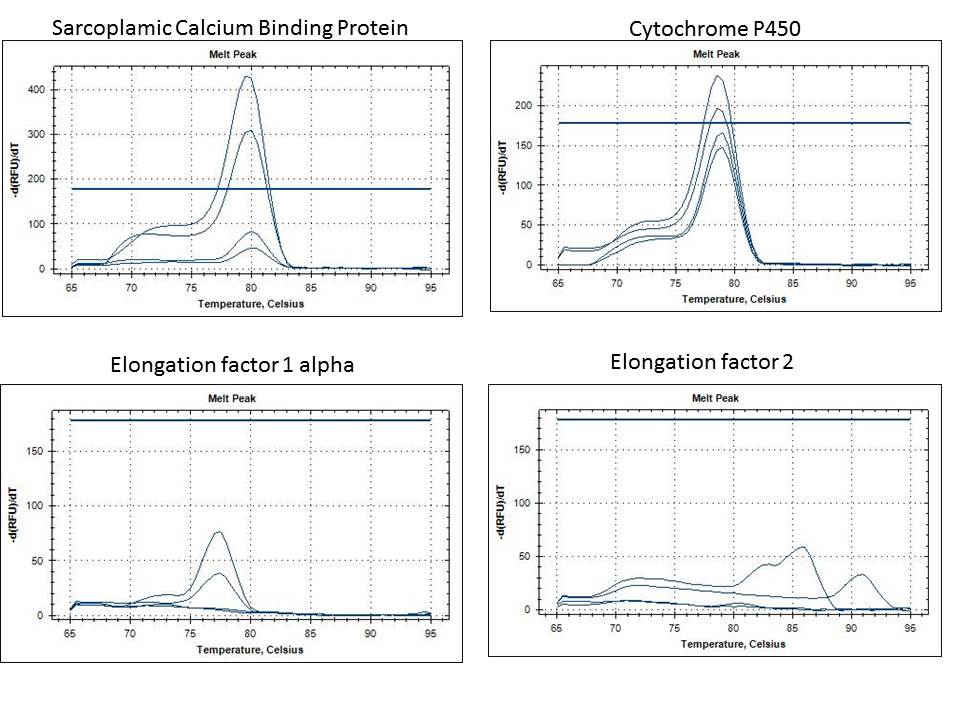
- ACP5 is not a typo, there just wasn't any ACP15 available, so this was a substituted primer.
Two bands were recovered. The bands in agarose were frozen at -20 degrees C. This time, there was a gene that was upregulated by the T-Iso group, possibly a fat storage gene if the copepods are not receiving a deficient diet?
July 14, 2009
After 2 more incubation at 55 degrees C for 10 minutes and an addition of 7.5 uL more of the DI H2o (0.1% DEPC), I was able to get consistent readings:
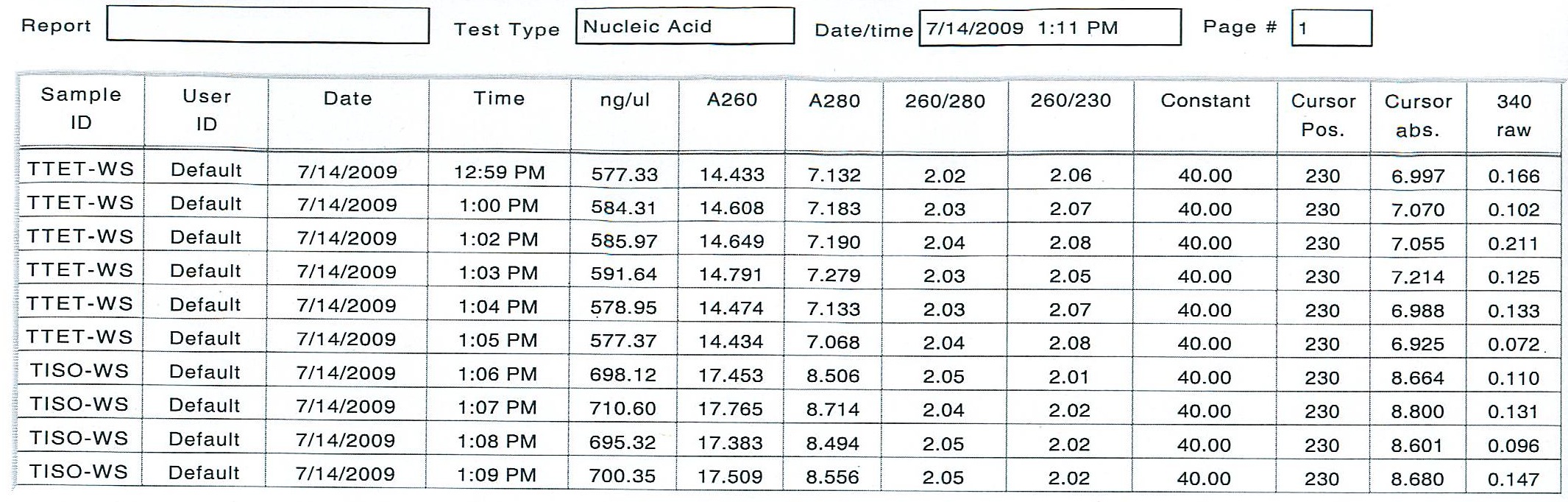
The average amount of RNA in the TTET working solution was 583.64 ng/uL and the average amount of RNA in the TISO working solution was 701.10 ng uL. I prepared the cDNA reactions according the Genefish instructions
Unfortunately, I switched the amounts for the basic RNA, so the reactions were not started with the same amount of RNA.
This is what I actually added:
TIso 1 = 5.14 uL RNA + 2.36 uL DIH20 + 2 uL dNTP: starting RNA = 3.60 ug RNA
TIso2 = 2.57 uL RNA + 4.93 uL DIH20 + 2 uL dNTP: starting RNA = 1.80 ug RNA
Tet1 = 4.27 uL RNA + 3.23 uL DIH20 + 2 uL dNTP: starting RNA = 2.49 ug RNA
Tet2 = 2.135 uL RNA + 5.36 uL DIH20 + 2 uL dNTP: starting RNA = 1.25 ug RNA
I will return to the stock solutions T-TISO2 and T-TET2 tomorrow and set up the cDNA reaction in the morning. It seems that an additional incubation of the stock in the heating block assists in smoothing out the RNA concentration estimates.
I have 25 uL of stock RNA left for both of these samples. (Original stock plus 10 uL dilution from this morning with DEPC (0.1%) in water to ensure no RNASE activity). There should be about 45 ug of RNA in the remaining stock solution, with a concentraiton of 1.8 ug/uL.
I took 5 uL (approximately 9 ug of RNA) from each and brought up the working solution volume to 18 uL, spec and placed the new working solution (TI-WS-2 and TT-WS-2) in the heating block at 55 degrees C for 20 minutes.
Differential Display
Just for kicks, used the cDNA produced from this first experiment to set up a differential display on the Tet and T-Iso. It was possible to figure out the ratio of T-ISO RNA to T-Tet was 1.45:1 before producing the cDNA, so this ratio was used to set up the differential display experiments.
A7 through A10 and the dT-ACP-2 were used to set up four reactions on the T-ISO, T-TET, and Control (DI-H20)
| T-ISO |
T-TET |
DIH2O |
|
| cDNA |
2 uL |
2.9 uL |
0 uL |
| arbitraty ACP 9 #7 through #10 |
2 uL |
2 uL |
2 uL |
| 10 uM dT-ACP-2 |
1 uL |
1 uL |
1 uL |
| 2 x SeeAMP ACP Master Mix |
10 uL |
10 uL |
10 uL |
| DIH2O |
5 uL |
4.1 uL |
7 uL |
| TOTAL |
20 uL |
20 uL |
20 uL |
| Segment |
|
Temp (degrees C) |
Duration (minutes) |
| 1 |
1 |
94 |
5 |
| 2 |
1 |
50 |
3 |
| 3 |
1 |
72 |
1 |
| 4 |
40 |
94 |
40 sec |
| 65 |
40 sec |
||
| 72 |
40 sec |
||
| 5 |
1 |
72 |
5 |
July 13, 2009
Today, I started the copepod feeding experiment on freshly-collected Tigriopus californicus specimens by taking previously starved animals (24 hours) and placing the in either Isochrysis (DHA-rich) or Tetraselmis (DHA-poor). The animals were fed in natural light for approximately 3 hours, then screened, placed on ice, rinsed with nanopure water and concentrated into snap-cap tubes for RNA extraction. To ensure sufficient extraction of RNA, I concentrated two groups of 200 animals each for each treatment (4 concentrated copeod samples).
I followed the same laboratory standard protocol for RNA extraction until I had a final volume of 20 uL for each of the four samples and here are the raw spec results:
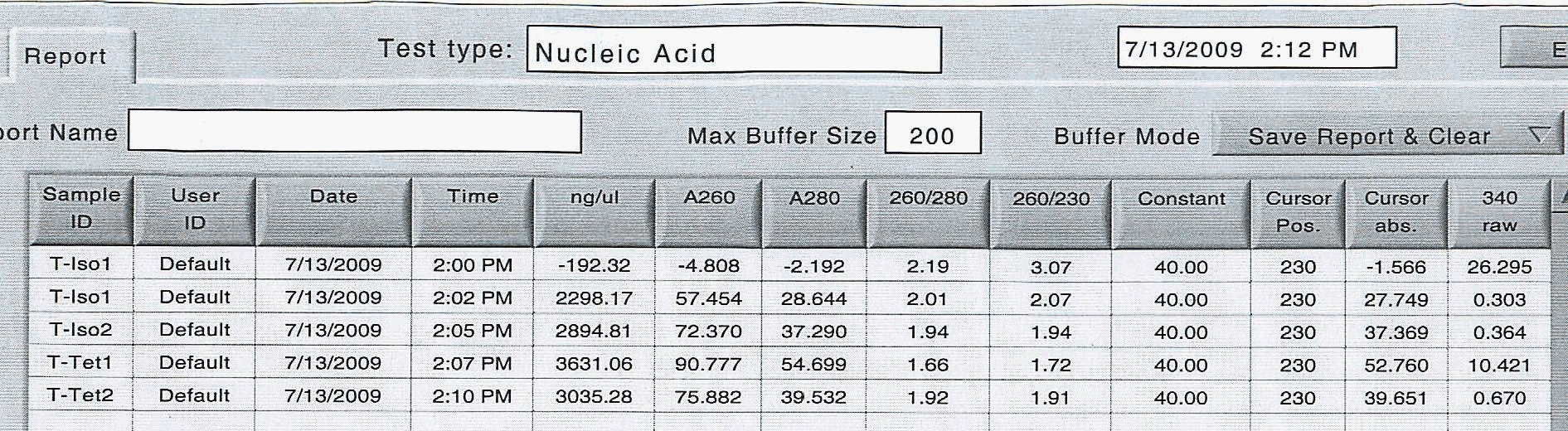 |
| Results of RNA extraction - 1st reading had an air bubble |
The very first reading had an air bubble.
T-Iso2 and T-Tet2 had similar quantities, so I will dilute these to become the working stock solution. I will use 3 uL of each sample, and bring up to a total volume of 18 uL each (an approximate concentration of 0.5 ug/uL) and then spec to find a final volume for the cDNA production.
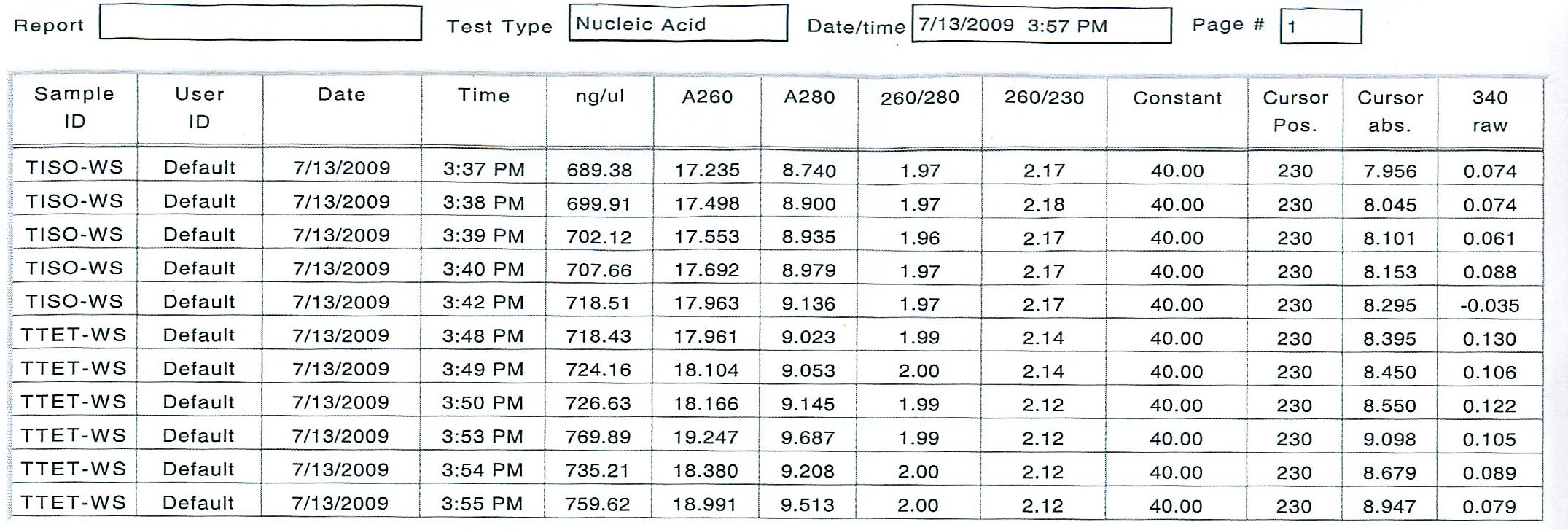
To see if all RNA was in solution, an additional 2 uL of stock RNA from TISO-2 and TTET-2 were added to the existing working solution and 10 uL of DIH20 (0.1% DEPC) was added. The new working solutions were incubated at 10 minutes at 55 degrees C, but the new numbers were not satisfactor as far as consistency.
July 9 & 10, 2009
RNA Precipitation
On Thursday, the two samples of RNA retrieved from the Tigriopus fed two different diets were precipitated.
50 uL RNA sample from Tigriopus treatments
1/10 volume ( 5 uL) 3 M Sodium Acetate pH 5.2
2x volume (110 uL) 100 % Ethanol
Incubated overnight at - 20 degrees C
Friday:
Sample spun at highest setting in 4 degrees C microcentrifuge for 30 minutes
Supernatant removed
1 mL 70% ethanol added
Sample spun at max speed in 4 degrees C for 10 minutes,
supernatant removed, spun again briefly, rest of ethanol drawn off
Once all ethanol removed, the RNA was resuspended in 10 uL of DI H2O (0.1% DEPC)
As it can be seen from the RNA concentration, some RNA was lost in the TI sample. Because it is easy enough to set up a second copeopd experiment with more animals, I am going to re-extract the RNA on Monday, and prepare a more concentrated sample the first time.
From 100 animals, it was possible to get at least 7 to 11 micrograms of material to work with. Since the protocol requires at least 3 micrograms, 100 animals should be sufficient. However, I think I will double up the experiment in case there are any problems with RNA extraction on this next round.
July 9, 2009
I have not found any significant matches in NCBI. I went through the .abi files using the program "Sequence Viewer" provided by ABI, and was able to make some guesses on assignments. However, more information did not improve the results. The stickler is that the putative sequences found in my Tigriopus samples last year using the same PCR procedure were not replicated this time. Curious.
This alignment is very sketchy also, but I did find it interesting that the Calanus sequence (C1) shared some alignment with the Acartia sequence(A1 and A2), but this may be due to random chance.
Ugh. Just realized I made a mistake when sending the sequences off for analysis ... I included both forward and reverse primers in the sample. So, they created two strands which overlapped. Well, I guess it is always good to learn from mistakes.
July 1, 2009
Results of the 8 sequences were returned today. They were very sketchy, leading me to believe that the bands captured by the delta-6 generic primer designed from plant/animal sequences was too generic perhaps. I am downloading a free .abi viewer in order to analyze the chromatograms.
Here are the sequences:
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNTCANTNNTNNNNNNNNNCNNNNNNTCCTGNGNCNNCNNTCANNNNCCNNNGA
NTCCAANCANNCGAAAANANCTTNNNTTCCTCCTCCTCCCTCATCTTCNGNTCCGNCNTAATGNNNNNNTGCTGCTGNNNNNTG
NAATNTATGGCTGNNTGNCANTCAGCTGCCTGCCTGTTACGCTCAGCCATATCACCCGCGCGCTTGCTCTCCGCCTCCTGCACCT
GGCTGTCCAACAGCCTCTCGTTCTNNNGNNGCTCCTTCTCCNNGTTCTCCTCCATGTGATCAANCNTCCTCTGCCTCTGNNTGTC
CTTNNANCACTANN
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNTNNNNNNNNNNNNANNANNGNNNNANNNNNNNNNNNNNNNNN
NNNNTANNNNNTNNNNCNAANNTAGGAAANNGNCGANTANGNGANNANNTCGGNANANTNNNAGNANNNNNTNNGNAGNT
GANNNNNNNNNTNCAGNGNNNNACGTGNNNCNNCNCATCGGGNCGNANNTNNNNTNNNNGNCNNNNNNNNNCNNGNCCNT
CNTNNNNNNNNN
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNGCTNNNNNNNNNNNNNNNNNNNNNNGTCNGTGNNNNNNGNNCNCCCNGN
CNNCNNNNNNGANCTNNNNCNNGNNNNNNGNCCNNNNNNNNTAANNNNN
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCTNNNNNNNNNNNNNNNNNTGANNNGNNNNNNANNTTGNNNNNNNNN
NNNNNNNNNNNNGNTCANNNNNNNNNNNNNNNNNCNNTATAANNCCGAGNNGANANNTTCGNNCNNGCCTNNNTANNNNG
NCTGCTGGNNCGTNTGNCNNNANNNTTNCGCANNTNNNNNNTNNTGNNNNGNTTGNNANTGNNNNNGCNCTGNTGNNANN
NTNNNNNNNNNNCNNNNNNNNNNNNNNNNNNNNCNNNNGN
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCATNNCNANNANCNNCNNNCCACTTGNNNTNNNGNNCANATTGNCTCT
CATATCANCATCCTCNNGTACCNATTGGACNANANNNNNTACTCCCTGACTGGGGTANACATTGTCACTATCAACCCTGA
TGCTGAACTGTCCATGCNNNGCNACAACCCNNNANGTCACTATGNNNNNNNTNANGGNNNNANATANGNANGNNNNNNNG
NNNNNNNNNNNNGANNNNNNNANNNNTNNNGNANNNGGTCANCCNTATGNNCNACNTCNCCNNGTNNNNGNACNTGNCTG
ANNCTGNNNANATNNNNNACCNNNTGNTCATA
NNNNNNNNNNNNNNNNNNNNNANTATCNCTGAGCATNTCTATCACCACCTCCACNNCTTGCGCTAAAGTCCACATTGTCT
CTCATATCAGCATCCTCATGTACAGATTGGACAAGACCAGCTACTCCCTGACTGGGGTAGACATTGTCACTATCAACCCT
GATGCTGAACTGTCCATGCTAAGCTACAACNCCAGAGGTCACTATGGNAGGGGTGAGGGGGGNANNTNTGGAGAAATAAC
TGTCTCCAAACATTTGATCAGCAGAGGGCTTGGCAACAAGGTCAGCCATNNGNACGACATCTCCCAGTACCTGTACGTGC
CTGACACTGAGCAGATCNANCACNACTTNNNCANN
NNNNNNNNNNNNNNNNNNNNNANNNNNNNNNNNNNNNNANCNNNNNNNCCANTTGNTCTGCAGTNCANNTTNNCNCNANA
TNNNCNNNNNNNNGTNNNNNNTNNNNNNNANGNNNTANTNNCTGNNNGGGTTANACNTTGNCACNANCANCCCNGATGCN
NNNNTGNCNNTGNNNANNNNNANCCCCNNANGTCACTANGNNNNGNNGNANNNNNNNNNNNANNNNNNNNNANNNNNNNN
ANNNNTNNANNNNNNCANNNNTNGNGNANANGGTCATCNNTNTGTACAANATCNNCNNGTNNNNGNAANNGCNTGNNNNT
GNNCANATCNNNNNCCNNNNNNNCATANN
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNTNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNTTNNNNGNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
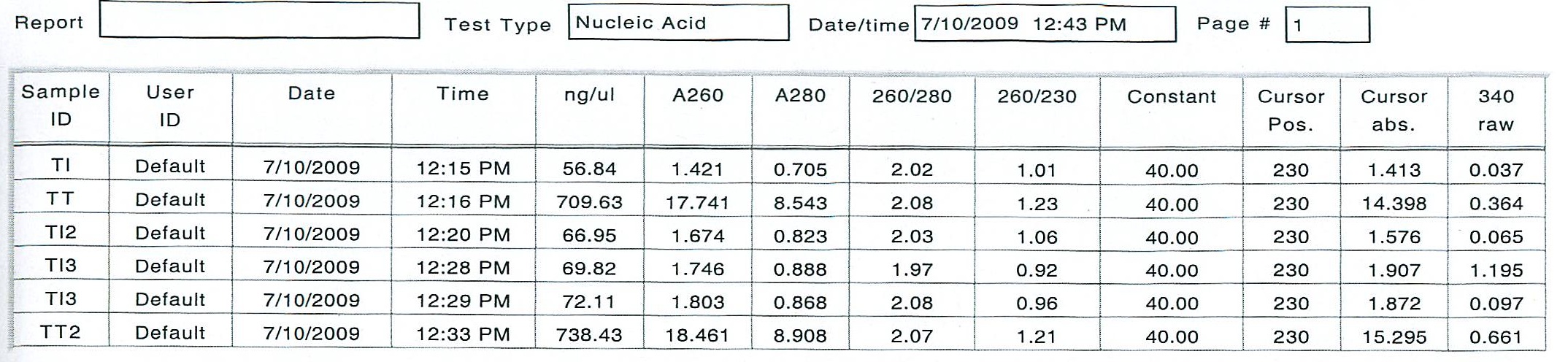
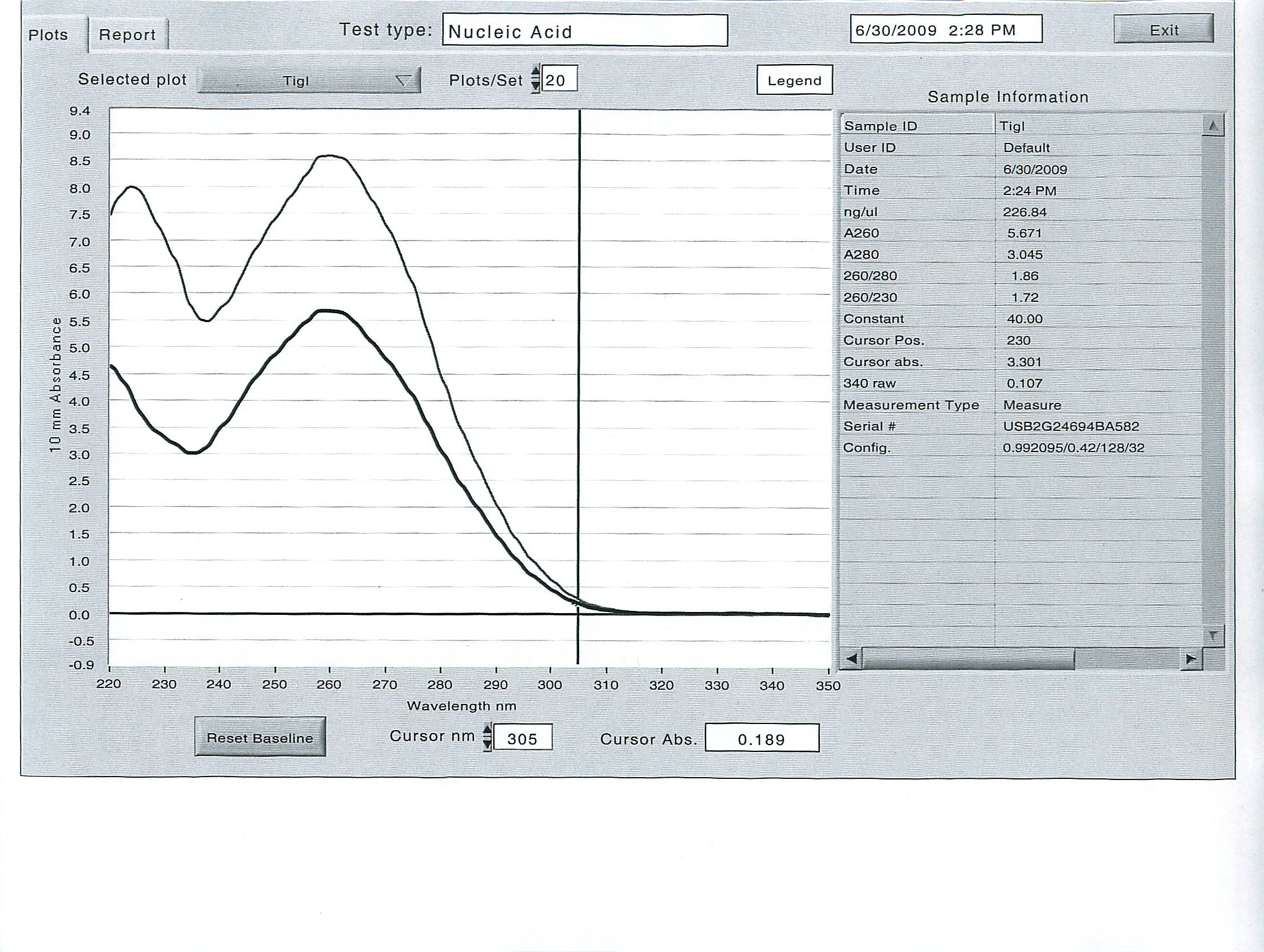
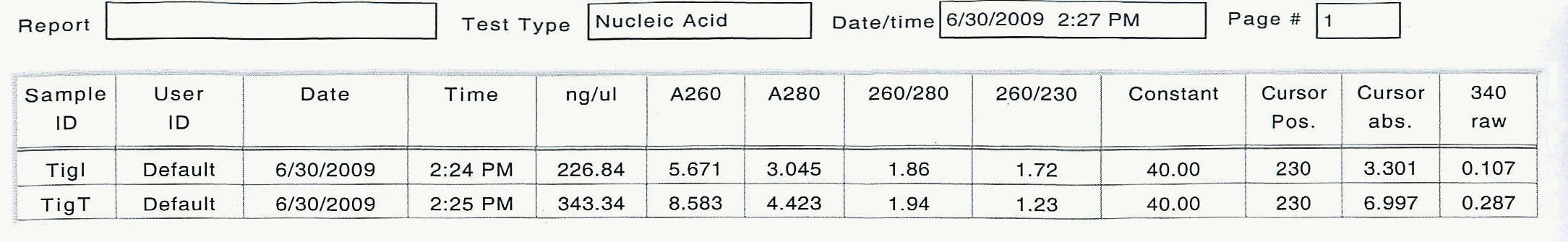
June 30, 2009
Tigriopus californicus experiment:
In an effort to get the copepods to ramp up their production of delta-6 desaturase, two diets were provided to two separate groups of 200 animals each, in excess: Isochrysis galbana - a brown algae with all essential fatty acids, and Tetraselmis suecica - a green prasinophyte deficient in the essential fatty acid DHA, which requires the use of delta-6 desaturase to produce by the animal. The copepods were kept on these diets for several hours, then screened, separated from the diets, rinsed and kept on ice. 100 animals each were counted into the 2 mL tubes, then homogenized and the total RNA was extracted with tri-reagent according to standard operating protocol.
Here are the results of the total RNA extraction from the spec on the final product at 50 uL volume:
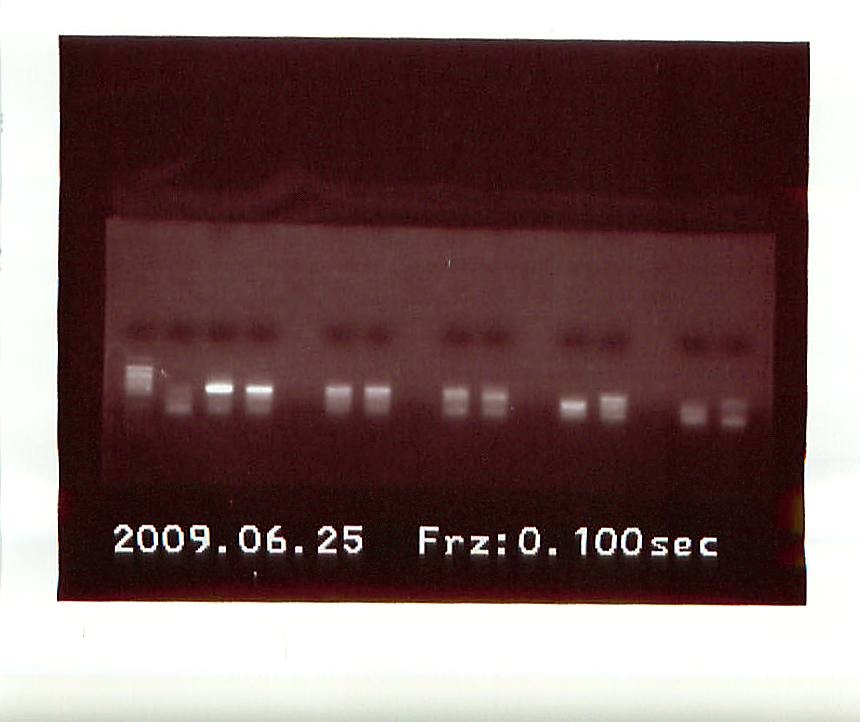
June 25, 2009
Another gel was run on the new PCR products.
 |
| 2nd PCR results |
From left to right, the gel had: Control, Acartia1, Acartia 2, empty, Calanus 1, Calanus 2, empty, Tisbe 1, Tisbe 2, empty, Tigriopus bottom band from Tigriopus 1 on first gel, Tigriopus top band from Tigriopus 1 on 2nd gel, empty, Tigriopus 2, Control.
Several band were highly fluorescent, so we took a gamble and sent off 8 DNA sequences to ASU:
Acartia 1&2, Calanus 1&2, Tisbe 1&2, and Tigriopus Top Sequence and Tigriopus Bottom Sequence.
June 24, 2009
PCR Product using Delta-6 Desaturase Primer with 4 species of copepods (DNA ladder and one control on the left, species in duplicate from left to right are Acartia tonsa, Calanus pacificus, Tisbe biminiensis, and Tigriopus californicus. One more control on the right completes the gel. )
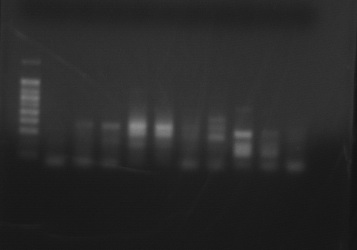 |
| Various species of copepods run with Delta-6 desaturase generic primer |
The PCR products were cut from this gel, and the DNA was extracted. A second PCR reaction was set up for the following bands:
Acartia tonsa 1&2 (3rd and 4th from the left), Calanus pacificus 1&2 (5th and 6th from the left), Tisbe 1&2 (7th and 8th from left) and Tigriopus 1 top band and bottom band (9th from left).
Annealing temperature was set to 50 degrees C, 50 ul of product was produced.
Hello Roberts Lab!
I am a visiting postdoc and I work on copepods. Copepods are the most numerous organism in the ocean and an important part of the food chain. In the past, I have worked on a variety of projects to capitalize on the unique nutritional properties of copepods for feeding fish larvae in aquaculture operations.
I am interested in determining the putative lipid conversion pathways used by copepods. Very little information is available about this diverse group of organisms (>12,000 species identified to date) on the NCBI genetic databases. Most information relates to the parasitic copepods that are a nuisance for salmon farms and other open ocean aquaculture operations. Copepods are called "sea lice", and they have single-handedly destroyed millions of dollars of fish operations in the past decade. There is a lot of interest in developing vaccines for the fish to prevent the attachment of the copepods. While the copepods themselves do not kill the fish, the holes they create in the fish flesh allow for the entry of harmful pathogens.
My work is not related to disease, but to to the basic physiology of the copepods. Some copepods are better than others at converting and storing lipids for the longterm and short term. While many researchers have speculated on the specific pathways of lipid conversion and storage, no definitive evidence provides a clear pathway for this important physiological mechanism in the copepods.
I am hoping to chip away a the pathway this summer by looking for the enzymes that might regulate the pathways for lipid desaturation and elongation in marine copepods. I have a variety of samples to work with, so I hope to find some clues as to the capabilities of copepods to produce these enzymes.
Wish me luck!